Computer Surface Science Laboratory
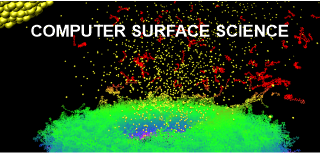
We use computer simulations to study surface processes at the atomic level, stimulated by impacts of energetic projectiles in the 3- (3D) and 2-dimensional (2D) systems. We are interested in modeling physicochemical processes occurring in thin metallic and semiconductor layers, organic nanostructures, as well as free-standing graphene and single and multiwall carbon nanotubes. Molecular Dynamics computer simulations are used to probe and visualize investigated phenomena.
Molecular Dynamics Computer Simulations
When a new phenomenon is observed, we usually want to understand what processes are responsible for it. Computer simulations are beneficial for achieving this goal as they allow the study of many properties that cannot be directly measured. For example, they allow to look inside the studied system and visually inspect the movement of atoms leading to a specific event or set of events. We can see what is happening there! In the MD approach, the temporal evolution of a given system is probed by integrating Hamilton’s equations of motion for all particles in the system. The forces among the atoms are described by a blend of empirical pair-wise additive and many-body potential energy functions.
We perform preliminary calculations on our computing server equipped with 356 computing units. The actual calculations are performed on computers belonging to the PLGrid supercomputer network.
Research Objectives

The rapid progress of miniaturization, which has taken place in recent years, has sparked a great deal of interest in studying physical and chemical phenomena occurring in devices with dimensions not much larger than the dimensions of individual atoms. This interest resulted in a quest for experimental techniques that would make it possible to measure the chemical composition on this scale. Examples of such techniques are Secondary Ion Mass Spectrometry (SIMS) and Secondary Neutral Mass Spectrometry (SNMS). These techniques use keV-energy projectiles to throw the particles residing at the analyzed surfaces into the vacuum. This phenomenon is called "sputtering". The emitted particles are then studied with mass spectrometers. The chemical composition of the analyzed surfaces is determined from the obtained mass spectra. In systems equipped with a scanned and focused ion beam, it is possible to additionally determine the location of a given compound in the sample volume. SIMS/SNMS techniques are so sensitive that they can find a single atom or molecule hidden among billions of other particles.

SIMS and SNMS have numerous practical applications. They are used in the design and control of integrated circuits or OLEDs that drive our electronic devices. They are also used to determine the age of geological structures, perform forgery checks of paintings and sculptures, or help to catch criminals based on evidence found at a crime scene. However, the most fascinating application of SIMS/SNMS is its use in medical research and diagnostics. For example, these techniques create three-dimensional chemical images of biological cells. They play a vital role in the development of new drugs. Compact and fully automatic SIMS systems are introduced into hospital operating rooms, where they support surgical procedures in the rapid identification of malignant tumor tissues. Currently, such identification can take days, and if the diagnosis is inauspicious, the patient must go through surgery again.
The primary objective of our research is to provide a theoretical background, explanation, and verification of recent findings and hypotheses, which emerge, as SIMS and SNMS expand beyond their current frontiers. State of the art computer simulations are used to study physicochemical processes emerging in new analytical configurations based on cluster projectiles and 2-dimensional and liquid supports. Research on liquid supports is particularly essential, as water is the main component of our body. It is crucial, therefore, to understand what is happening in such an environment.
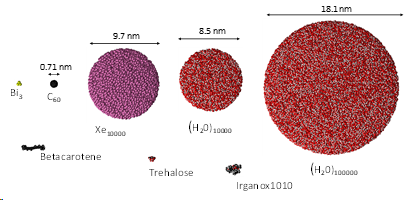
Our Research Projects
Cluster bombardments of solid surfaces
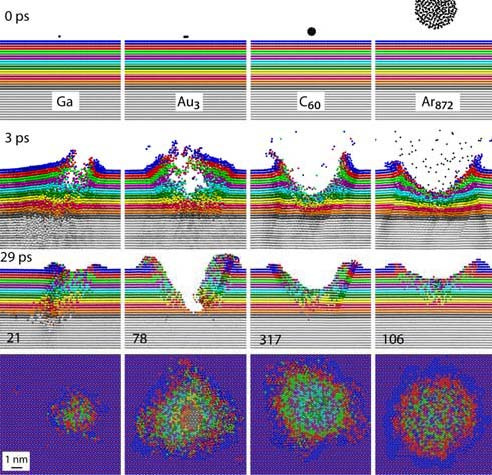
Cluster projectiles appeared in SIMS/SNMS techniques when they proved to allow for a significant increase in particle emission efficiency, combined with a lower probability of fragmentation of the emitted molecules. In the SIMS/SNMS techniques, the identification of the sampled material is achieved by analyzing the mass spectra of the emitted particles. Therefore, the spectrum must consist mainly of peaks corresponding to original molecules, not their fragments. The computer simulations made it possible to understand why cluster projectile bombardment is so different from the impacts of monatomic projectiles [1].
An impact of a monoatomic projectile initiates a cascade of collisions in the material. Individual atoms hit other atoms in a sophisticated game of atomic billiards. Initially, all kinetic energy is carried by a single atom. When such a projectile collides with a molecule, it breaks into pieces. Monoatomic projectiles are also small. They easily penetrate into the material. As a result, the primary kinetic energy is deposited deep in the sample, far from the surface. This energy is not available for the emission of particles from the surface. It only mixes the sample material and leads to damage inside the material. These processes change the original geometric structure and chemical composition of the sample, making it challenging to perform chemical analysis with high spatial resolution [1].
Cluster projectiles are much larger. As a result, they can not penetrate the material and deposit their energy close to the surface—the shallow deposition of energy results in a strong emission of particles from the material's surface. For example, a C60 projectile emits 15 times more material than a Ga projectile with the same kinetic energy. The emission of many particles leads to a formation of a crater on the bombarded surface, as shown on the left. Since the primary kinetic energy is partitioned among many atoms in a cluster, the energy carried by a single atom is small. Also, the motion of these atoms is spatially correlated. The impact of cluster projectile is more gentle than that of their atomic counterparts. As a result, it is possible now to uplift intact molecules from the surface. Therefore, cluster projectiles (e.g., C60 or Arn) are currently a workhorse of SIMS analysis organic and biological structures. However, new cluster projectiles are still on the lookout for increasing emissions efficiency. Interesting candidates currently being investigated are, for example, cluster projectiles made of H2O or CO2 molecules.
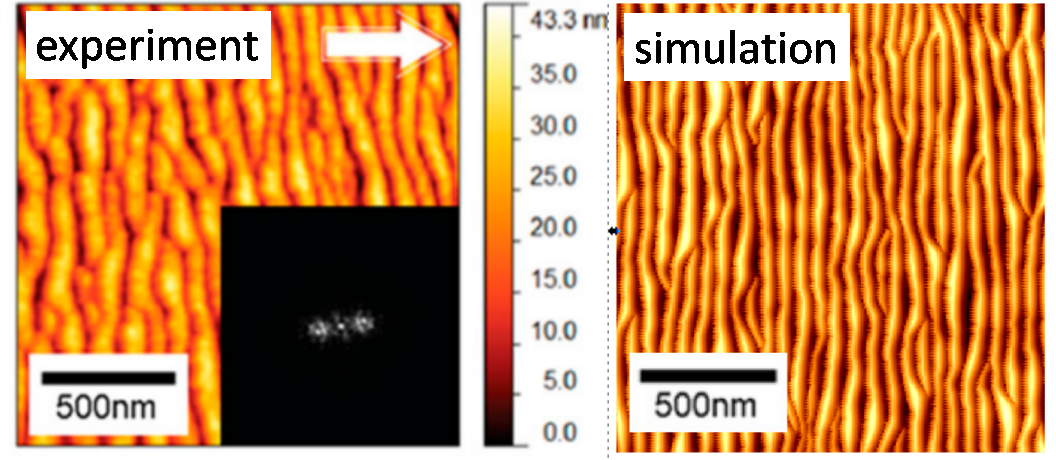
One of the most important factors influencing the spatial accuracy of the chemical composition measurement, i.e., depth resolution, is the appearance of the surface roughness analysis. One type of such roughness is a ripple. The appearance of ripples is a limiting factor for the depth resolution that can be obtained because the accuracy cannot be greater than the amplitude of the ripples. Sometimes this amplitude can be huge. The problem of ripple formation has been extensively studied for monatomic projectiles. However, still little is known about this process when cluster projectiles bombard the surface. We have recently proposed a simple model that allows for an intuitive explanation of the phenomenon of ripple formation through mass transfer caused by a cluster projectile impact [2]. The shape of this transfer dependence on the impact angle turned out to be of crucial importance. By combining this model with computer simulations, one can predict when ripples will appear and what to do to avoid them. Despite its simplicity, this model produces results that are quantitatively consistent with the results of experimental measurements. Our studies indicate that the problems resulting from ripples' appearance can be reduced by lowering the ion beam's angle relative to the normal to the surface or by increasing its kinetic energy.
Sputtering and modification of 2D systems (graphene, carbon nanotubes)
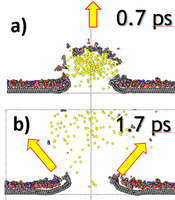
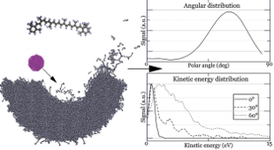
We focus on a theoretical study of processes taking place during the analysis of two-dimensional (2D) van der Waals materials by SIMS/SNMS. These systems have attracted worldwide attention due to their remarkable optical, mechanical, and electronic transport properties. Most of the attention has been focused on graphene as the principal material in future 2D electronics. However, materials like hexagonal boron nitride (h-BN) and molybdenum disulfide (h-MoS2) are also crucial as having similar structural properties as graphene; they differ both in electrical conductivity and elastic constants. The capability to visualize the chemical structures of two-dimensional materials and their interfaces is crucial for devising highly efficient ultrathin electronic devices. Our recent studies also show that two-dimensional systems used as substrates can increase SIMS detection efficiency. This finding potentially opens the door to the chemical analysis of small nano-objects, supramolecular ensembles, and attomole amounts of analyte detection [3, 4]. We show that organic material deposited on graphene is ejected by a gentle interaction with unfolding graphene substrate, as presented on the left.
Bombardment of liquid matrices with cluster projectiles
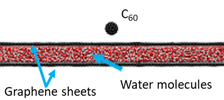
The chemical imaging of biological structures is currently one of the hottest topics in SIMS and SNMS. In principle, the most appropriate substrate to perform such studies is water, a natural environment for biosamples. However, such an approach has been hampered by problems associated with the incompatibility of high-vacuum conditions required for SIMS/SNMS operation and water samples. One way to overcome this limitation is to encapsulate investigated biostructures in a liquid confined between graphene membranes as presented on the left. Unfortunately, it has been found that graphene undergoes almost instant self-healing. As a result, minimal emission of the investigated analyte can be obtained from this system [5].
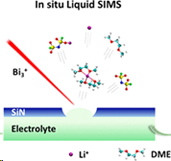
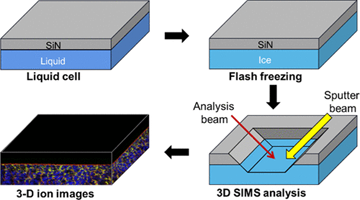
Another possibility is to encapsulate the investigated liquid in a small vacuum-compatible microfluid cell that is capped by a thin SiN membrane at the top, as shown in Figure on the left. After incorporating this cell into the analysis chamber, a small, approximately 2 μm hole is drilled by a focused ion beam. It has been shown that the device can still sustain a reasonably high vacuum (<5 × 10−7 mbar) in the instrument chamber. This system has already generated numerous fascinating and puzzling observations that require theoretical explanation. MS simulations can assist in this task [6].
Development of a fast ReaxFF potential
Apart from significant progress in the computer modeling of SIMS-related phenomena during the last several years, there is still a very limited number of organic systems, which can be studied by a full atomistic approach. This situation is caused by the necessity to engage much more sophisticated many-body potentials to describe interactions between atoms in organic samples. Such potentials are very time-expensive. While it is true that atomistic potentials capable of simulating chemical reactions could be found in the literature, most of them were too slow to be applied in SIMS-related studies. In these studies, millions of atoms are required to contain primary kinetic energy carried by a typical projectile used during SIMS analysis.
We proposed modifying the ReaxFF potential that eliminates the time-consuming charge equilibration, includes an expanded training set for condensed phases, and has a proper repulsive wall for energetic particle bombardment simulations. We have recently applied this approach to developing a fast force field for systems composed of carbon and hydrogen atoms [7] and carbon, hydrogen and oxygen atoms [8]. An example of research with a new potential is shown on the left. The influence of the incident angle on the efficiency of the emission of molecules from the β-carotene layers was investigated [9]. In these studies, it was shown that the new potential allowed to speed up the calculations several times without losing their accuracy.
[1] B.J. Garrison and Z. Postawa, a chapter in "ToF-SIMS - Surface Analysis by Mass Spectrometry - 2nd Edition (2013).
[2] D. Maciazek, M. Kanski, and Z. Postawa, Analytical Chemistry 92 (2020) 7349.
[3] S. Verkhoturov, M. Gołuński, D. Verkhoturov, B. Czerwinski, M. Eller, S.Geng, Z. Postawa, E.A. Schweikert, J. Chem. Phys. 150 (2019) 160901.;
[4] M. Golunski, S. Hrabar, and Z. Postawa, Applied Surface Science 539 (2021) 148259
[5] S. Verkhoturov, M. Gołuński, Z. Postawa, in preparation.
[6] W. Guo, M. Kanski, W. Liu, M. Gołuński, Y. Zhou, Y. Wang, C. Cheng, Y. Du, Z. Postawa, W.D. Wei, and Z. Zhua, Analytical Chemistry 92 (2020) 13785 - featured publication.
[7] M. Kanski, D. Maciazek, Z. Postawa, Ch.M. Ashraf, A.C.T. van Duin, B.J. Garrison, J. Phys. Chem. Lett. 9 (2018) 359.
[8] M. Kański, S. Hrabar, A.C.T. van Duin, Z. Postawa, J. Phys. Chem. Lett. 13 (2022) 628.
[9] M. Kański and Z. Postawa, Analytical Chemistry 91 (2019) 9161 - wyróżniona publikacja.
Group members
- Prof. Zbigniew Postawa - Head
- Dr. Michał Kański
- Dr. Soukaina Louerdi
- Mgr Sviatoslav Hrabar - PhD Student
Magazine covers illustrating the results of our research
render(['images' => [ [ 'file_path' => '/img/groups/comp-sci/winter_2010.jpg', 'alt' => '', 'caption' => '' ], [ 'file_path' => '/img/groups/comp-sci/cover.jpg', 'alt' => '', 'caption' => '' ], [ 'file_path' => '/img/groups/comp-sci/cov150h_zp.jpg', 'alt' => '', 'caption' => '' ], [ 'file_path' => '/img/groups/comp-sci/cover_SIMS.jpg', 'alt' => '', 'caption' => '' ], [ 'file_path' => '/img/groups/comp-sci/cover_ancham.jpg', 'alt' => '', 'caption' => '' ], [ 'file_path' => '/img/groups/comp-sci/cover_anchem_2020.jpg', 'alt' => '', 'caption' => '' ] ]]); ?>Collaborators
- Gerhard Hobler, TU Wien, Austria
- Emile Schweikert, The Texas A&M University, USA
- Stanislav Verkhoturov, The Texas A&M University, USA
- Nicholas Winograd, The Pennsylvania State University, USA
Contact
More can be found on the group website at the Group page.
 prof. dr hab. Zbigniew Postawa
prof. dr hab. Zbigniew Postawa